5.30 10.11
Covariance Matrix of Latent Variables
KQ | CH | DP | QH | |
-------- | -------- | -------- | -------- | |
KQ | 1.00 | |||
CH | 0.50 | 1.00 | ||
DP | 0.64 | 0.21 | 1.00 | |
QH | 0.68 | 0.31 | 0.49 | 1.00 |
Có thể bạn quan tâm!
-
 Phân Tích Nhân Tố Khám Phá Efa Của Biến Phụ Thuộc
Phân Tích Nhân Tố Khám Phá Efa Của Biến Phụ Thuộc -
 Nghiên cứu chuỗi cung ứng dịch vụ du lịch tại vùng Đồng bằng sông Hồng và Duyên hải Đông Bắc Việt Nam - 27
Nghiên cứu chuỗi cung ứng dịch vụ du lịch tại vùng Đồng bằng sông Hồng và Duyên hải Đông Bắc Việt Nam - 27 -
 Nghiên cứu chuỗi cung ứng dịch vụ du lịch tại vùng Đồng bằng sông Hồng và Duyên hải Đông Bắc Việt Nam - 28
Nghiên cứu chuỗi cung ứng dịch vụ du lịch tại vùng Đồng bằng sông Hồng và Duyên hải Đông Bắc Việt Nam - 28 -
 Nghiên cứu chuỗi cung ứng dịch vụ du lịch tại vùng Đồng bằng sông Hồng và Duyên hải Đông Bắc Việt Nam - 30
Nghiên cứu chuỗi cung ứng dịch vụ du lịch tại vùng Đồng bằng sông Hồng và Duyên hải Đông Bắc Việt Nam - 30
Xem toàn bộ 241 trang tài liệu này.
Goodness of Fit Statistics Degrees of Freedom = 203
Minimum Fit Function Chi-Square = 830.21 (P = 0.0) Normal Theory Weighted Least Squares Chi-Square = 901.26 (P = 0.0)
Estimated Non-centrality Parameter (NCP) = 898.26
90 Percent Confidence Interval for NCP = (592.86 ; 911.13)
Minimum Fit Function Value = 2.67 Population Discrepancy Function Value (F0) = 2.86
90 Percent Confidence Interval for F0 = (2.56 ; 3.18) Root Mean Square Error of Approximation (RMSEA) = 0.052
90 Percent Confidence Interval for RMSEA = (0.051 ; 0.053) P-Value for Test of Close Fit (RMSEA < 0.05) = 0.00
Expected Cross-Validation Index (ECVI) = 3.73
90 Percent Confidence Interval for ECVI = (3.43 ; 4.05) ECVI for Saturated Model = 1.45
ECVI for Independence Model = 25.92
Chi-Square for Independence Model with 231 Degrees of Freedom = 9003.75
Independence AIC = 9047.75 Model AIC = 1301.26
Saturated AIC = 506.00 Independence CAIC = 9154.63
Model CAIC = 1544.15
Saturated CAIC = 1735.06
Normed Fit Index (NFI) = 0.90 Non-Normed Fit Index (NNFI) = 0.91
Parsimony Normed Fit Index (PNFI) = 0.82 Comparative Fit Index (CFI) = 0.92 Incremental Fit Index (IFI) = 0.92
Relative Fit Index (RFI) = 0.88
Critical N (CN) = 95.84
Root Mean Square Residual (RMR) = 0.052 Standardized RMR = 0.078
Goodness of Fit Index (GFI) = 0.96 Adjusted Goodness of Fit Index (AGFI) = 0.80
Parsimony Goodness of Fit Index (PGFI) = 0.77 The Modification Indices Suggest to Add the
Path to from Decrease in Chi-Square New Estimate
DP4 CH 14.7 0.13
The Modification Indices Suggest to Add an Error Covariance Between and Decrease in Chi-Square New Estimate
KQ1 | 85.0 | 0.21 | |
KQ3 | KQ1 | 28.6 | 0.13 |
KQ3 | KQ2 | 13.6 | 0.09 |
KQ4 | KQ1 | 23.6 | -0.11 |
KQ4 | KQ3 | 21.4 | -0.12 |
KQ5 | KQ1 | 12.9 | -0.08 |
KQ5 | KQ3 | 24.9 | -0.12 |
KQ6 | KQ1 | 40.1 | -0.13 |
KQ6 | KQ2 | 34.5 | -0.12 |
KQ6 | KQ3 | 17.1 | -0.09 |
KQ6 | KQ4 | 63.3 | 0.17 |
KQ6 | KQ5 | 31.7 | 0.11 |
KQ7 | KQ1 | 18.3 | -0.10 |
KQ7 | KQ2 | 22.1 | -0.11 |
KQ7 | KQ4 | 15.7 | 0.09 |
KQ7 | KQ5 | 10.4 | 0.07 |
KQ7 | KQ6 | 35.1 | 0.12 |
KQ8 | KQ1 | 14.2 | -0.09 |
KQ8 | KQ2 | 18.2 | -0.11 |
KQ8 | KQ3 | 14.3 | -0.10 |
KQ8 | KQ4 | 15.4 | 0.10 |
KQ8 | KQ5 | 15.0 | 0.09 |
KQ8 | KQ6 | 23.2 | 0.11 |
KQ8 | KQ7 | 19.4 | 0.11 |
KQ9 | KQ1 | 14.8 | -0.10 |
KQ9 | KQ2 | 8.6 | -0.07 |
KQ9 | KQ5 | 13.1 | 0.09 |
KQ9 | KQ6 | 22.1 | 0.11 |
KQ9 | KQ8 | 34.3 | 0.16 |
DP1 | KQ1 | 10.7 | 0.06 |
DP1 | KQ3 | 8.4 | 0.06 |
DP1 | KQ4 | 19.2 | -0.08 |
DP2 | KQ1 | 9.0 | -0.04 |
DP2 | KQ5 | 8.6 | 0.04 |
DP4 | KQ1 | 28.4 | 0.10 |
DP4 | KQ3 | 43.9 | 0.14 |
DP4 | KQ6 | 21.5 | -0.08 |
DP4 | CH1 | 16.6 | 0.09 |
DP4 | DP2 | 17.7 | -0.06 |
QH1 | KQ3 | 13.7 | 0.09 |
QH1 | DP4 | 15.2 | 0.08 |
QH2 | QH1 | 24.3 | 0.12 |
QH3 | KQ4 | 8.4 | 0.06 |
QH3 | QH2 | 11.2 | 0.07 |
QH4 | QH1 | 10.8 | -0.07 |
QH4 | QH2 | 33.1 | -0.13 |
QH5 | QH3 | 16.4 | -0.08 |
QH5 | QH4 | 13.6 | 0.08 |
QH6 | QH5 | 8.6 | 0.06 |
9. MÔ HÌNH SEM 2
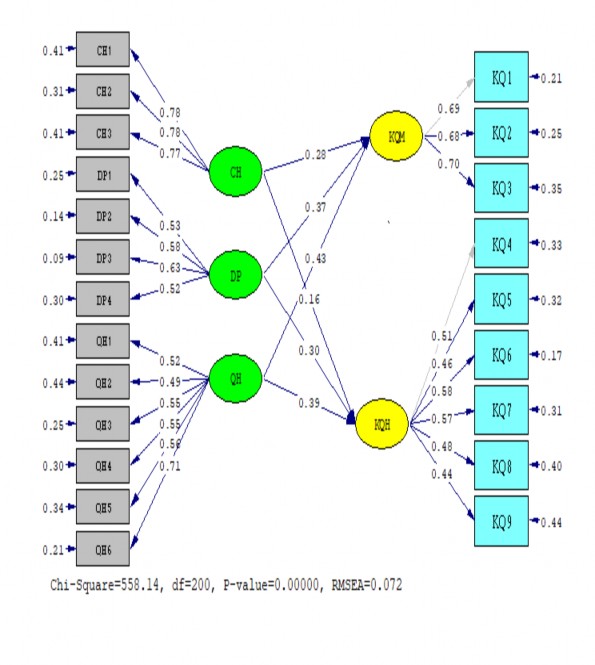
Sample Size = 350 PATH ANALYSIS
Covariance Matrix
KQ1 | KQ2 | KQ3 | KQ4 | KQ5 | ||
-------- | -------- | -------- | -------- | -------- | ----- | |
--- | ||||||
KQ1 | 0.69 | |||||
KQ2 | 0.49 | 0.71 | ||||
KQ3 | 0.47 | 0.45 | 0.84 | |||
KQ4 | 0.15 | 0.21 | 0.17 | 0.59 | ||
KQ5 | 0.16 | 0.20 | 0.16 | 0.20 | 0.53 | |
KQ6 | 0.16 | 0.18 | 0.22 | 0.32 | 0.27 | |
0.50 | ||||||
KQ7 | 0.23 | 0.23 | 0.31 | 0.29 | 0.26 | |
0.32 | ||||||
KQ8 | 0.14 | 0.14 | 0.16 | 0.24 | 0.23 | |
0.26 | ||||||
KQ9 | 0.13 | 0.16 | 0.17 | 0.19 | 0.22 | |
0.25 | ||||||
CH1 | 0.37 | 0.32 | 0.40 | 0.08 | 0.07 | |
0.08 | ||||||
CH2 | 0.32 | 0.27 | 0.36 | 0.04 | 0.10 | |
0.10 | ||||||
CH3 | 0.30 | 0.26 | 0.36 | 0.08 | 0.08 | |
0.05 | ||||||
DP1 | 0.25 | 0.21 | 0.29 | 0.07 | 0.12 | |
0.12 | ||||||
DP2 | 0.20 | 0.25 | 0.25 | 0.17 | 0.18 | |
0.17 | ||||||
DP3 | 0.22 | 0.26 | 0.26 | 0.18 | 0.14 | |
0.18 | ||||||
DP4 | 0.31 | 0.25 | 0.38 | 0.12 | 0.13 | |
0.10 | ||||||
QH1 | 0.24 | 0.25 | 0.32 | 0.10 | 0.14 | |
0.13 | ||||||
QH2 | 0.21 | 0.19 | 0.21 | 0.15 | 0.11 | |
0.16 | ||||||
QH3 | 0.26 | 0.27 | 0.23 | 0.21 | 0.13 | |
0.21 | ||||||
QH4 | 0.21 | 0.24 | 0.27 | 0.19 | 0.16 | |
0.20 | ||||||
QH5 | 0.24 | 0.21 | 0.27 | 0.11 | 0.19 | |
0.18 | ||||||
QH6 | 0.26 | 0.26 | 0.30 | 0.16 | 0.17 | |
0.18 |
Covariance Matrix
CH3
---
KQ7 KQ8 KQ9 CH1 CH2
-------- -------- -------- -------- -------- ----- KQ7 0.64
KQ8 | 0.29 0.63 | |||||
KQ9 | 0.24 0.28 | 0.63 | ||||
CH1 | 0.16 0.10 | 0.11 | 1.02 | |||
CH2 | 0.13 0.11 | 0.11 | 0.60 | 0.91 | ||
0.99 | CH3 DP1 | 0.16 0.08 0.18 0.11 | 0.12 0.09 | 0.59 0.08 | 0.61 0.06 | |
0.06 | DP2 | 0.19 0.11 | 0.14 | 0.11 | 0.09 | |
0.06 | DP3 | 0.16 0.10 | 0.12 | 0.12 | 0.08 | |
0.10 | DP4 | 0.15 0.08 | 0.11 | 0.25 | 0.14 | |
0.14 | QH1 | 0.20 0.06 | 0.07 | 0.17 | 0.12 | |
0.14 | QH2 | 0.16 0.07 | 0.07 | 0.12 | 0.12 | |
0.10 | QH3 | 0.24 0.13 | 0.13 | 0.13 | 0.15 | |
0.13 | QH4 | 0.23 0.16 | 0.12 | 0.13 | 0.15 | |
0.08 | QH5 | 0.18 0.13 | 0.07 | 0.20 | 0.16 | |
0.10 | QH6 | 0.22 0.15 | 0.10 | 0.20 | 0.16 | |
0.10 | ||||||
Covariance Matrix | ||||||
QH2 --- | DP1 | DP1 DP2 -------- -------- 0.53 | DP3 -------- | DP4 -------- | QH1 -------- | ----- |
DP2 | 0.30 0.48 | |||||
DP3 | 0.33 0.37 | 0.49 | ||||
DP4 | 0.29 0.26 | 0.33 | 0.57 | |||
QH1 | 0.14 0.14 | 0.14 | 0.21 | 0.68 | ||
0.68 | QH2 QH3 | 0.12 0.14 0.13 0.19 | 0.14 0.18 | 0.10 0.13 | 0.35 0.31 | |
0.32 | QH4 | 0.15 0.18 | 0.19 | 0.16 | 0.23 | |
0.17 | QH5 | 0.17 0.15 | 0.18 | 0.17 | 0.26 | |
0.24 | QH6 | 0.17 0.21 | 0.18 | 0.16 | 0.36 | |
0.35 | ||||||
Covariance Matrix | ||||||
QH3 | QH3 QH4 -------- -------- 0.55 | QH5 -------- | QH6 -------- | |||
QH4 | 0.29 0.60 | |||||
QH5 | 0.26 0.36 | 0.65 | ||||
QH6 | 0.38 0.41 | 0.43 | 0.71 |
PATH ANALYSIS
Number of Iterations = 11
LISREL Estimates (Maximum Likelihood) Measurement Equations
KQ1 = 0.69*KQM, Errorvar.= 0.21 , R² = 0.69
(0.024)
8.86
KQ2 = 0.68*KQM, Errorvar.= 0.25 , R² = 0.65 (0.042) (0.026)
16.26 9.63
KQ3 = 0.70*KQM, Errorvar.= 0.35 , R² = 0.59 (0.046) (0.033)
15.33 10.48
KQ4 = 0.51*KQH, Errorvar.= 0.33 , R² = 0.44
(0.029)
11.42
KQ5 = 0.46*KQH, Errorvar.= 0.32 , R² = 0.40 (0.045) (0.027)
10.13 11.73
KQ6 = 0.58*KQH, Errorvar.= 0.17 , R² = 0.66 (0.047) (0.019)
12.36 8.76
KQ7 = 0.57*KQH, Errorvar.= 0.31 , R² = 0.51 (0.051) (0.029)
11.27 10.81
KQ8 = 0.48*KQH, Errorvar.= 0.40 , R² = 0.36 (0.049) (0.034)
9.75 11.93
KQ9 = 0.44*KQH, Errorvar.= 0.44 , R² = 0.30 (0.049) (0.036)
9.02 12.23
CH1 = 0.78*CH, Errorvar.= 0.41 , R² = 0.60 (0.049) (0.044)
15.75 9.30
CH2 = 0.78*CH, Errorvar.= 0.31 , R² = 0.66 (0.046) (0.038)
16.87 8.02
CH3 = 0.77*CH, Errorvar.= 0.41 , R² = 0.59 (0.049) (0.043)
15.68 9.38
DP1 = 0.53*DP, Errorvar.= 0.25 , R² = 0.52 (0.035) (0.022)
15.12 11.61
DP2 = 0.58*DP, Errorvar.= 0.14 , R² = 0.70 (0.031) (0.015)
18.52 9.61
DP3 = 0.63*DP, Errorvar.= 0.086 , R² = 0.82 (0.030) (0.013)
21.06 6.42
DP4 = 0.52*DP, Errorvar.= 0.30 , R² = 0.47 (0.037) (0.025)
14.08 11.93
QH1 = 0.52*QH, Errorvar.= 0.41 , R² = 0.40 (0.042) (0.034)
12.42 12.04
QH2 = 0.49*QH, Errorvar.= 0.44 , R² = 0.35 (0.042) (0.036)
11.44 12.26
QH3 = 0.55*QH, Errorvar.= 0.25 , R² = 0.55 (0.036) (0.022)
15.52 10.95
QH4 = 0.55*QH, Errorvar.= 0.30 , R² = 0.50 (0.038) (0.026)
14.47 11.40
QH5 = 0.56*QH, Errorvar.= 0.34 , R² = 0.48 (0.040) (0.029)
14.15 11.52
QH6 = 0.71*QH, Errorvar.= 0.21 , R² = 0.70 (0.038) (0.024)
18.39 8.97
Structural Equations
KQM = 0.43*QH + 0.37*DP + 0.28*CH, Errorvar.= 0.32 , R² = 0.68 (0.021) (0.033) (0.045) (0.039)
8.52 7.00 5.55 6.49
KQH = 0.16*CH + 0.30*DP + 0.39*QH, Errorvar.= 0.64 , R² = 0.56
(0.035) (0.021) | (0.020) | ||||
1.08 | 4.28 5.52 | 6.27 | |||
Correlation Matrix of Independent Variables CH DP QH -------- -------- -------- CH 1.00 DP 0.22 1.00 (0.06) 3.62 QH 0.31 0.49 1.00 (0.06) (0.05) 5.31 10.13 Covariance Matrix of Latent Variables | |||||
KQM | KQM -------- 1.00 | KQH -------- | CH -------- | DP -------- | QH -------- |
KQH | 0.45 | 1.00 | |||
CH | 0.61 | 0.24 | 1.00 | ||
DP | 0.61 | 0.48 | 0.22 | 1.00 | |
QH | 0.62 | 0.54 | 0.31 | 0.49 | 1.00 |
Goodness of Fit Statistics
Degrees of Freedom = 200
Minimum Fit Function Chi-Square = 570.92 (P = 0.0) Normal Theory Weighted Least Squares Chi-Square = 558.14 (P =
0.0)
Estimated Non-centrality Parameter (NCP) = 358.14
90 Percent Confidence Interval for NCP = (291.51 ; 432.42)
Minimum Fit Function Value = 1.64 Population Discrepancy Function Value (F0) = 1.03
90 Percent Confidence Interval for F0 = (0.84 ; 1.24) Root Mean Square Error of Approximation (RMSEA) = 0.072
90 Percent Confidence Interval for RMSEA = (0.065 ; 0.079) P-Value for Test of Close Fit (RMSEA < 0.05) = 0.00
Expected Cross-Validation Index (ECVI) = 1.90
90 Percent Confidence Interval for ECVI = (1.71 ; 2.12) ECVI for Saturated Model = 1.45
ECVI for Independence Model = 25.92
Chi-Square for Independence Model with 231 Degrees of Freedom = 9003.75
Independence AIC = 9047.75 Model AIC = 664.14
Saturated AIC = 506.00