RMSEA
RMSEA | LO 90 | HI 90 | PCLOSE | |
Default model | ,049 | ,042 | ,056 | ,600 |
Independence model | ,198 | ,193 | ,203 | ,000 |
Có thể bạn quan tâm!
-
 Nhóm Các Yếu Tố Tác Động Đến Kết Quả Kinh Doanh
Nhóm Các Yếu Tố Tác Động Đến Kết Quả Kinh Doanh -
 = “Hoàn Toàn Không Đồng Ý”, 2 = “Rất Không Đồng Ý”, 3 = “Không Đồng Ý”, 4 = “Phân Vân”, 5 = “Đồng Ý”, 6 = “Rất Đồng Ý” Và 7 = “Hoàn Toàn
= “Hoàn Toàn Không Đồng Ý”, 2 = “Rất Không Đồng Ý”, 3 = “Không Đồng Ý”, 4 = “Phân Vân”, 5 = “Đồng Ý”, 6 = “Rất Đồng Ý” Và 7 = “Hoàn Toàn -
 Tác động của vốn xã hội đối với kết quả kinh doanh của doanh nghiệp - trường hợp nghiên cứu ngành Dệt may khu vực phía Nam, Việt Nam - 32
Tác động của vốn xã hội đối với kết quả kinh doanh của doanh nghiệp - trường hợp nghiên cứu ngành Dệt may khu vực phía Nam, Việt Nam - 32 -
 Kết Quả Phân Tích Độ Tin Cậy Thang Đo Chính Thức
Kết Quả Phân Tích Độ Tin Cậy Thang Đo Chính Thức -
 Tác động của vốn xã hội đối với kết quả kinh doanh của doanh nghiệp - trường hợp nghiên cứu ngành Dệt may khu vực phía Nam, Việt Nam - 35
Tác động của vốn xã hội đối với kết quả kinh doanh của doanh nghiệp - trường hợp nghiên cứu ngành Dệt may khu vực phía Nam, Việt Nam - 35 -
 Tác động của vốn xã hội đối với kết quả kinh doanh của doanh nghiệp - trường hợp nghiên cứu ngành Dệt may khu vực phía Nam, Việt Nam - 36
Tác động của vốn xã hội đối với kết quả kinh doanh của doanh nghiệp - trường hợp nghiên cứu ngành Dệt may khu vực phía Nam, Việt Nam - 36
Xem toàn bộ 295 trang tài liệu này.
AIC
AIC | BCC | BIC | CAIC | |
Default model | 762,807 | 781,128 | 1057,221 | 1137,221 |
Saturated model | 870,000 | 969,618 | 2470,875 | 2905,875 |
Independence model | 5125,125 | 5131,766 | 5231,850 | 5260,850 |
ECVI
ECVI | LO 90 | HI 90 | MECVI | |
Default model | 2,612 | 2,393 | 2,858 | 2,675 |
Saturated model | 2,979 | 2,979 | 2,979 | 3,321 |
Independence model | 17,552 | 16,777 | 18,349 | 17,575 |
HOELTER
HOELTER HOELTER .05 .01 | |
Default model Independence model | 194 204 27 28 |
CBSEM standardized
8.3. KIỂM ĐỊNH MÔ HÌNH LÝ THUYẾT BẰNG CBSEM
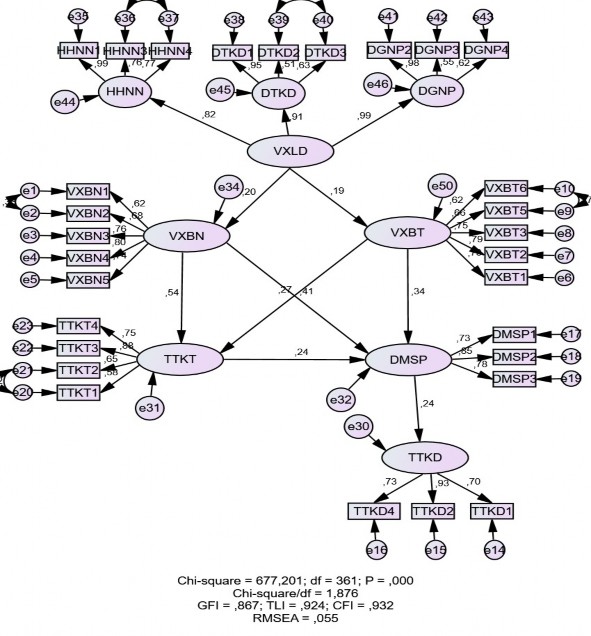
Scalar Estimates (Group number 1 Default model) Maximum Likelihood Estimates
Regression Weights: (Group number 1 Default model)
Estimate | S.E. | C.R. | P Label | |||
VXBN | < | VXLD | ,150 | ,049 | 3,080 | ,002 |
VXBT | < | VXLD | ,109 | ,037 | 2,941 | ,003 |
TTKT | < | VXBN | ,421 | ,063 | 6,634 | *** |
TTKT | < | VXBT | ,283 | ,070 | 4,028 | *** |
DMSP | < | VXBN | ,449 | ,086 | 5,221 | *** |
DMSP | < | VXBT | ,498 | ,100 | 4,987 | *** |
DMSP | < | TTKT | ,331 | ,112 | 2,948 | ,003 |
HHNN | < | VXLD | ,922 | ,048 | 19,308 | *** |
DTKD | < | VXLD | 1,048 | ,048 | 21,949 | *** |
DGNP | < | VXLD | 1,000 | |||
TTKD | < | DMSP | ,251 | ,073 | 3,454 | *** |
VXBN5 | < | VXBN | 1,000 | |||
VXBN4 | < | VXBN | 1,069 | ,084 | 12,653 | *** |
VXBN3 | < | VXBN | 1,113 | ,092 | 12,115 | *** |
VXBN2 | < | VXBN | ,900 | ,083 | 10,790 | *** |
VXBN1 | < | VXBN | ,895 | ,092 | 9,768 | *** |
VXBT6 | < | VXBT | 1,000 | |||
VXBT5 | < | VXBT | 1,059 | ,100 | 10,552 | *** |
VXBT3 | < | VXBT | 1,420 | ,146 | 9,754 | *** |
VXBT2 | < | VXBT | 1,435 | ,143 | 10,004 | *** |
VXBT1 | < | VXBT | 1,456 | ,152 | 9,581 | *** |
TTKD2 | < | TTKD | 1,518 | ,129 | 11,764 | *** |
DMSP1 | < | DMSP | 1,000 | |||
DMSP2 | < | DMSP | 1,123 | ,087 | 12,888 | *** |
DMSP3 | < | DMSP | 1,041 | ,085 | 12,192 | *** |
TTKT1 | < | TTKT | 1,000 | |||
TTKT2 | < | TTKT | 1,109 | ,099 | 11,176 | *** |
TTKT3 | < | TTKT | 1,522 | ,157 | 9,694 | *** |
TTKT4 | < | TTKT | 1,498 | ,162 | 9,271 | *** |
HHNN1 | < | HHNN | 1,000 | |||
HHNN3 | < | HHNN | ,716 | ,043 | 16,637 | *** |
HHNN4 | < | HHNN | ,722 | ,043 | 16,983 | *** |
DTKD1 | < | DTKD | 1,000 | |||
DTKD2 | < | DTKD | ,470 | ,050 | 9,310 | *** |
Estimate | S.E. | C.R. | P Label | |||
DTKD3 | < | DTKD | ,533 | ,044 | 12,082 | *** |
DGNP2 | < | DGNP | 1,000 | |||
DGNP3 | < | DGNP | ,477 | ,045 | 10,608 | *** |
DGNP4 | < | DGNP | ,558 | ,045 | 12,505 | *** |
TTKD4 | < | TTKD | 1,067 | ,092 | 11,612 | *** |
TTKD1 | < | TTKD | 1,000 |
Standardized Regression Weights: (Group number 1 Default model)
Estimate | |||
VXBN | < | VXLD | ,199 |
VXBT | < | VXLD | ,195 |
TTKT | < | VXBN | ,543 |
TTKT | < | VXBT | ,272 |
DMSP | < | VXBN | ,411 |
DMSP | < | VXBT | ,341 |
DMSP | < | TTKT | ,235 |
HHNN | < | VXLD | ,816 |
DTKD | < | VXLD | ,912 |
DGNP | < | VXLD | ,989 |
TTKD | < | DMSP | ,237 |
VXBN5 | < | VXBN | ,743 |
VXBN4 | < | VXBN | ,798 |
VXBN3 | < | VXBN | ,759 |
VXBN2 | < | VXBN | ,677 |
VXBN1 | < | VXBN | ,615 |
VXBT6 | < | VXBT | ,619 |
VXBT5 | < | VXBT | ,657 |
VXBT3 | < | VXBT | ,754 |
VXBT2 | < | VXBT | ,789 |
VXBT1 | < | VXBT | ,732 |
TTKD2 | < | TTKD | ,932 |
DMSP1 | < | DMSP | ,728 |
DMSP2 | < | DMSP | ,852 |
DMSP3 | < | DMSP | ,779 |
TTKT1 | < | TTKT | ,581 |
TTKT2 | < | TTKT | ,646 |
TTKT3 | < | TTKT | ,878 |
TTKT4 | < | TTKT | ,747 |
HHNN1 | < | HHNN | ,991 |
HHNN3 | < | HHNN | ,762 |
Estimate | |||
HHNN4 | < | HHNN | ,772 |
DTKD1 | < | DTKD | ,948 |
DTKD2 | < | DTKD | ,514 |
DTKD3 | < | DTKD | ,631 |
DGNP2 | < | DGNP | ,978 |
DGNP3 | < | DGNP | ,551 |
DGNP4 | < | DGNP | ,622 |
TTKD4 | < | TTKD | ,732 |
TTKD1 | < | TTKD | ,705 |
Covariances: (Group number 1 Default model)
Estimate | S.E. | C.R. | P Label | |||
e2 | <> | e1 | ,208 | ,046 | 4,497 | *** |
e10 | <> | e9 | ,132 | ,035 | 3,726 | *** |
e39 | <> | e40 | ,115 | ,043 | 2,695 | ,007 |
e36 | <> | e37 | ,224 | ,037 | 6,012 | *** |
e20 | <> | e21 | ,264 | ,046 | 5,717 | *** |
Correlations: (Group number 1 Default model)
Estimate | |||
e2 | <> | e1 | ,328 |
e10 | <> | e9 | ,271 |
e39 | <> | e40 | ,169 |
e36 | <> | e37 | ,484 |
e20 | <> | e21 | ,422 |
Variances: (Group number 1 Default model)
Estimate | S.E. | C.R. | P Label | |
VXLD | 1,002 | ,093 | 10,811 | *** |
e34 | ,542 | ,078 | 6,928 | *** |
e50 | ,303 | ,056 | 5,378 | *** |
e31 | ,211 | ,044 | 4,806 | *** |
e32 | ,334 | ,056 | 6,001 | *** |
e30 | ,713 | ,112 | 6,374 | *** |
e44 | ,427 | ,060 | 7,131 | *** |
e45 | ,222 | ,063 | 3,546 | *** |
e46 | ,022 | ,039 | ,558 | ,577 |
e5 | ,459 | ,048 | 9,570 | *** |
Estimate | S.E. | C.R. | P Label | |
e4 | ,369 | ,043 | 8,493 | *** |
e3 | ,514 | ,055 | 9,295 | *** |
e2 | ,542 | ,053 | 10,293 | *** |
e1 | ,743 | ,069 | 10,747 | *** |
e10 | ,508 | ,048 | 10,554 | *** |
e9 | ,465 | ,045 | 10,255 | *** |
e8 | ,484 | ,053 | 9,072 | *** |
e7 | ,393 | ,047 | 8,299 | *** |
e6 | ,578 | ,061 | 9,446 | *** |
e14 | ,767 | ,079 | 9,742 | *** |
e15 | ,264 | ,108 | 2,433 | ,015 |
e16 | ,747 | ,081 | 9,185 | *** |
e17 | ,598 | ,061 | 9,744 | *** |
e18 | ,320 | ,049 | 6,581 | *** |
e19 | ,472 | ,054 | 8,775 | *** |
e20 | ,668 | ,061 | 10,921 | *** |
e21 | ,584 | ,056 | 10,468 | *** |
e22 | ,234 | ,047 | 4,989 | *** |
e23 | ,604 | ,067 | 9,075 | *** |
e35 | ,022 | ,043 | ,528 | ,597 |
e36 | ,473 | ,045 | 10,501 | *** |
e37 | ,452 | ,044 | 10,353 | *** |
e38 | ,150 | ,054 | 2,751 | ,006 |
e39 | ,811 | ,069 | 11,712 | *** |
e40 | ,569 | ,050 | 11,373 | *** |
e41 | ,047 | ,031 | 1,553 | ,121 |
e42 | ,533 | ,045 | 11,832 | *** |
e43 | ,506 | ,043 | 11,684 | *** |
Model | NPAR | CMIN | DF | P | CMIN/DF |
Default model | 74 | 677,201 | 361 | ,000 | 1,876 |
Saturated model | 435 | ,000 | 0 | ||
Independence model | 29 | 5067,125 | 406 | ,000 | 12,481 |
Model Fit Summary CMIN
RMR, GFI
Model | RMR | GFI | AGFI | PGFI |
RMR | GFI | AGFI | PGFI | |
Saturated model | ,000 | 1,000 | ||
Independence model | ,341 | ,281 | ,230 | ,262 |
Baseline Comparisons
NFI Delta1 | RFI rho1 | IFI Delta2 | TLI rho2 | CFI | |
Default model | ,866 | ,850 | ,933 | ,924 | ,932 |
Saturated model | 1,000 | 1,000 | 1,000 | ||
Independence model | ,000 | ,000 | ,000 | ,000 | ,000 |
ParsimonyAdjusted Measures
PRATIO | PNFI | PCFI | |
Default model | ,889 | ,770 | ,829 |
Saturated model | ,000 | ,000 | ,000 |
Independence model | 1,000 | ,000 | ,000 |
NCP
NCP | LO 90 | HI 90 | |
Default model | 316,201 | 246,759 | 393,457 |
Saturated model | ,000 | ,000 | ,000 |
Independence model | 4661,125 | 4434,930 | 4893,775 |
FMIN
FMIN | F0 | LO 90 | HI 90 | |
Default model | 2,319 | 1,083 | ,845 | 1,347 |
Saturated model | ,000 | ,000 | ,000 | ,000 |
Independence model | 17,353 | 15,963 | 15,188 | 16,760 |
RMSEA
RMSEA | LO 90 | HI 90 | PCLOSE | |
Default model | ,055 | ,048 | ,061 | ,108 |
Independence model | ,198 | ,193 | ,203 | ,000 |
AIC
AIC | BCC | BIC | CAIC | |
Default model | 825,201 | 842,147 | 1097,533 | 1171,533 |
Saturated model | 870,000 | 969,618 | 2470,875 | 2905,875 |
Independence model | 5125,125 | 5131,766 | 5231,850 | 5260,850 |
ECVI
ECVI | LO 90 | HI 90 | MECVI | |
Default model | 2,826 | 2,588 | 3,091 | 2,884 |
Saturated model | 2,979 | 2,979 | 2,979 | 3,321 |
Independence model | 17,552 | 16,777 | 18,349 | 17,575 |
HOELTER
HOELTER HOELTER .05 .01 | |
Default model Independence model | 176 184 27 28 |
8.4. KIỂM ĐỊNH BOOTSTRAP
Regression Weights: (Group number 1 Default model)
SE SESE | Mean | Bias | SEBias | ||||
VXBN | < | VXLD | .057 | .002 | .148 | .002 | .003 |
VXBT | < | VXLD | .049 | .002 | .107 | .002 | .002 |
TTKT | < | VXBN | .097 | .003 | .427 | .006 | .004 |
TTKT | < | VXBT | .107 | .003 | .285 | .002 | .005 |
DMSP | < | VXBN | .109 | .003 | .453 | .003 | .005 |
DMSP | < | VXBT | .152 | .005 | .500 | .002 | .007 |
DMSP | < | TTKT | .149 | .005 | .350 | .018 | .007 |
HHNN | < | VXLD | .054 | .002 | .924 | .002 | .002 |
DTKD | < | VXLD | .056 | .002 | 1.049 | .001 | .003 |
DGNP | < | VXLD | .000 | .000 | 1.000 | .000 | .000 |
TTKD | < | DMSP | .089 | .003 | .247 | .005 | .004 |
VXBN5 | < | VXBN | .000 | .000 | 1.000 | .000 | .000 |
VXBN4 | < | VXBN | .118 | .004 | 1.079 | .011 | .005 |
VXBN3 | < | VXBN | .122 | .004 | 1.124 | .011 | .005 |
VXBN2 | < | VXBN | .099 | .003 | .907 | .007 | .004 |
VXBN1 | < | VXBN | .092 | .003 | .896 | .001 | .004 |
VXBT6 | < | VXBT | .000 | .000 | 1.000 | .000 | .000 |
VXBT5 | < | VXBT | .106 | .003 | 1.065 | .006 | .005 |
VXBT3 | < | VXBT | .160 | .005 | 1.438 | .018 | .007 |
VXBT2 | < | VXBT | .164 | .005 | 1.451 | .016 | .007 |
VXBT1 | < | VXBT | .171 | .005 | 1.472 | .016 | .008 |
TTKD2 | < | TTKD | .137 | .004 | 1.534 | .016 | .006 |
DMSP1 | < | DMSP | .000 | .000 | 1.000 | .000 | .000 |
DMSP2 | < | DMSP | .085 | .003 | 1.127 | .004 | .004 |
DMSP3 | < | DMSP | .103 | .003 | 1.045 | .004 | .005 |