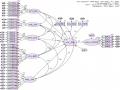
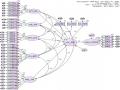
Maximum Likelihood Estimates
Regression Weights: (Group number 1 - Default model)
Estimate | S.E. C.R. | P | Label | ||
CL_DD | <--- | MT_DL | .159 | .010 16.066 | *** |
CL_DD | <--- | CS_HT | .210 | .012 17.488 | *** |
CL_DD | <--- | GIA_CA | .146 | .012 12.074 | *** |
CL_DD | <--- | VH_BD | .293 | .018 16.611 | *** |
CL_DD | <--- | HD_TN | .178 | .013 13.375 | *** |
HAI_LONG | <--- | CL_DD | 1.184 | .069 17.081 | *** |
MTDL6 | <--- | MT_DL | .959 | .048 20.048 | *** |
MTDL5 | <--- | MT_DL | .979 | .049 19.924 | *** |
MTDL4 | <--- | MT_DL | 1.000 | ||
MTDL3 | <--- | MT_DL | 1.042 | .053 19.774 | *** |
MTDL2 | <--- | MT_DL | .824 | .044 18.880 | *** |
MTDL1 | <--- | MT_DL | .811 | .045 18.124 | *** |
CSHT4 | <--- | CS_HT | 1.000 | ||
CSHT3 | <--- | CS_HT | 1.042 | .046 22.569 | *** |
CSHT2 | <--- | CS_HT | 1.169 | .048 24.515 | *** |
CSHT1 | <--- | CS_HT | .766 | .039 19.555 | *** |
GC5 | <--- | GIA_CA | 1.000 | ||
GC4 | <--- | GIA_CA | 1.186 | .078 15.119 | *** |
GC3 | <--- | GIA_CA | 1.292 | .081 16.019 | *** |
GC2 | <--- | GIA_CA | 1.189 | .078 15.206 | *** |
GC1 | <--- | GIA_CA | 1.174 | .075 15.707 | *** |
VHBD9 | <--- | VH_BD | 1.000 | ||
VHBD8 | <--- | VH_BD | 1.223 | .069 17.706 | *** |
VHBD7 | <--- | VH_BD | 1.320 | .077 17.113 | *** |
Có thể bạn quan tâm!
-
 Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 19
Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 19 -
 Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 20
Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 20 -
 Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 21
Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 21 -
 Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 23
Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 23 -
 Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 24
Nghiên cứu các yếu tố ảnh hưởng đến sự hài lòng của khách du lịch cộng đồng vùng Tây Bắc - 24
Xem toàn bộ 199 trang tài liệu này.

Estimate | S.E. C.R. | P | Label | ||
VHBD6 | <--- | VH_BD | 1.193 | .075 15.966 | *** |
VHBD5 | <--- | VH_BD | 1.321 | .075 17.581 | *** |
VHBD4 | <--- | VH_BD | 1.251 | .069 18.118 | *** |
VHBD3 | <--- | VH_BD | 1.064 | .063 16.858 | *** |
VHBD2 | <--- | VH_BD | 1.028 | .058 17.678 | *** |
VHBD1 | <--- | VH_BD | 1.238 | .069 17.915 | *** |
HDTN7 | <--- | HD_TN | 1.000 | ||
HDTN6 | <--- | HD_TN | 1.019 | .051 19.981 | *** |
HDTN5 | <--- | HD_TN | .938 | .054 17.356 | *** |
HDTN4 | <--- | HD_TN | .925 | .048 19.178 | *** |
HDTN3 | <--- | HD_TN | .827 | .046 18.068 | *** |
HDTN2 | <--- | HD_TN | .946 | .050 18.825 | *** |
HDTN1 | <--- | HD_TN | .948 | .048 19.777 | *** |
SA1 | <--- | HAI_LONG | 1.000 | ||
SA2 | <--- | HAI_LONG | .946 | .051 18.476 | *** |
SA3 | <--- | HAI_LONG | .997 | .052 19.018 | *** |
SA4 | <--- | HAI_LONG | 1.026 | .050 20.530 | *** |
SA5 | <--- | HAI_LONG | .896 | .050 18.096 | *** |
CLDD1 | <--- | CL_DD | 1.000 | ||
CLDD2 | <--- | CL_DD | 1.084 | .059 18.231 | *** |
CLDD3 | <--- | CL_DD | 1.112 | .061 18.118 | *** |
Standardized Regression Weights: (Group number 1 - Default model)
Estimate | |||
CL_DD | <--- | MT_DL | .455 |
CL_DD | <--- | CS_HT | .488 |
CL_DD | <--- | GIA_CA | .305 |
CL_DD | <--- | VH_BD | .686 |
CL_DD | <--- | HD_TN | .299 |
HAI_LONG | <--- | CL_DD | .893 |
MTDL6 | <--- | MT_DL | .799 |
MTDL5 | <--- | MT_DL | .795 |
MTDL4 | <--- | MT_DL | .782 |
MTDL3 | <--- | MT_DL | .790 |
MTDL2 | <--- | MT_DL | .761 |
MTDL1 | <--- | MT_DL | .736 |
CSHT4 | <--- | CS_HT | .842 |
CSHT3 | <--- | CS_HT | .813 |
CSHT2 | <--- | CS_HT | .859 |
CSHT1 | <--- | CS_HT | .737 |
GC5 | <--- | GIA_CA | .672 |
GC4 | <--- | GIA_CA | .745 |
GC3 | <--- | GIA_CA | .800 |
GC2 | <--- | GIA_CA | .750 |
GC1 | <--- | GIA_CA | .780 |
VHBD9 | <--- | VH_BD | .721 |
VHBD8 | <--- | VH_BD | .767 |
VHBD7 | <--- | VH_BD | .742 |
Estimate | |||
VHBD6 | <--- | VH_BD | .694 |
VHBD5 | <--- | VH_BD | .762 |
VHBD4 | <--- | VH_BD | .784 |
VHBD3 | <--- | VH_BD | .731 |
VHBD2 | <--- | VH_BD | .766 |
VHBD1 | <--- | VH_BD | .776 |
HDTN7 | <--- | HD_TN | .794 |
HDTN6 | <--- | HD_TN | .788 |
HDTN5 | <--- | HD_TN | .704 |
HDTN4 | <--- | HD_TN | .763 |
HDTN3 | <--- | HD_TN | .727 |
HDTN2 | <--- | HD_TN | .751 |
HDTN1 | <--- | HD_TN | .781 |
SA1 | <--- | HAI_LONG | .784 |
SA2 | <--- | HAI_LONG | .742 |
SA3 | <--- | HAI_LONG | .760 |
SA4 | <--- | HAI_LONG | .808 |
SA5 | <--- | HAI_LONG | .730 |
CLDD1 | <--- | CL_DD | .714 |
CLDD2 | <--- | CL_DD | .754 |
CLDD3 | <--- | CL_DD | .749 |
Covariances: (Group number 1 - Default model)
Estimate | S.E. | C.R. | P | Label | |
MT_DL <--> CS_HT | -.121 | .084 | -1.444 | .149 | |
MT_DL <--> GIA_CA | .164 | .077 | 2.134 | .033 | |
MT_DL <--> VH_BD | -.008 | .083 | -.091 | .927 | |
MT_DL <--> HD_TN | .085 | .060 | 1.417 | .157 | |
CS_HT <--> GIA_CA | -.047 | .062 | -.757 | .449 | |
CS_HT <--> VH_BD | -.040 | .068 | -.592 | .554 | |
CS_HT <--> HD_TN | .124 | .050 | 2.509 | .012 | |
GIA_CA <--> VH_BD | .006 | .062 | .092 | .927 | |
GIA_CA <--> HD_TN | .083 | .045 | 1.835 | .066 | |
VH_BD <--> HD_TN | .026 | .048 | .545 | .586 | |
Correlations: (Group number 1 - Default model)
Estimate | |||
MT_DL | <--> | CS_HT | -.069 |
MT_DL | <--> | GIA_CA | .104 |
MT_DL | <--> | VH_BD | -.004 |
MT_DL | <--> | HD_TN | .067 |
CS_HT | <--> | GIA_CA | -.037 |
CS_HT | <--> | VH_BD | -.028 |
CS_HT | <--> | HD_TN | .121 |
GIA_CA | <--> | VH_BD | .004 |
GIA_CA | <--> | HD_TN | .089 |
VH_BD | <--> | HD_TN | .026 |
Variances: (Group number 1 - Default model)
Estimate | S.E. C.R. P | Label | |
MT_DL | 2.141 | .202 10.610 *** | |
CS_HT | 1.426 | .120 11.841 *** | |
GIA_CA | 1.150 | .136 8.444 *** | |
VH_BD | 1.439 | .150 9.574 *** | |
HD_TN | .741 | .068 10.832 *** | |
e41 | -.038 | .004 -9.745 *** | |
e37 | .093 | .012 8.022 *** | |
e1 | 1.114 | .080 13.877 *** | |
e2 | 1.194 | .086 13.948 *** | |
e3 | 1.362 | .096 14.167 *** | |
e4 | 1.398 | .100 14.031 *** | |
e5 | 1.057 | .073 14.460 *** | |
e6 | 1.192 | .081 14.751 *** | |
e7 | .585 | .046 12.636 *** | |
e8 | .795 | .059 13.424 *** | |
e9 | .693 | .058 12.036 *** | |
e10 | .705 | .048 14.644 *** | |
e11 | 1.396 | .095 14.671 *** | |
e12 | 1.300 | .095 13.722 *** | |
e13 | 1.082 | .086 12.526 *** | |
e14 | 1.265 | .093 13.633 *** | |
e15 | 1.019 | .078 13.021 *** | |
e16 | 1.329 | .086 15.437 *** | |
e17 | 1.507 | .100 15.097 *** | |
Estimate | S.E. C.R. P | Label | |
e18 | 2.046 | .134 15.297 *** | |
e19 | 2.205 | .141 15.589 *** | |
e20 | 1.818 | .120 15.143 *** | |
e21 | 1.410 | .094 14.931 *** | |
e22 | 1.416 | .092 15.372 *** | |
e23 | 1.072 | .071 15.108 *** | |
e24 | 1.461 | .097 15.016 *** | |
e25 | .434 | .031 13.799 *** | |
e26 | .470 | .034 13.914 *** | |
e27 | .665 | .044 14.953 *** | |
e28 | .456 | .032 14.303 *** | |
e29 | .452 | .031 14.730 *** | |
e30 | .511 | .035 14.451 *** | |
e31 | .424 | .030 14.021 *** | |
e32 | .290 | .020 14.287 *** | |
e33 | .338 | .023 14.812 *** | |
e34 | .337 | .023 14.614 *** | |
e35 | .260 | .019 13.875 *** | |
e36 | .326 | .022 14.935 *** | |
e38 | .253 | .015 16.798 *** | |
e39 | .235 | .014 16.709 *** | |
e40 | .254 | .015 16.724 *** | |
Squared Multiple Correlations: (Group number 1 - Default model)
Estimate | |
CL_DD | 1.146 |
HAI_LONG | .798 |
CLDD3 | .562 |
CLDD2 | .568 |
CLDD1 | .510 |
SA5 | .532 |
SA4 | .652 |
SA3 | .577 |
SA2 | .551 |
SA1 | .614 |
HDTN1 | .611 |
HDTN2 | .565 |
HDTN3 | .529 |
HDTN4 | .582 |
HDTN5 | .495 |
HDTN6 | .621 |
HDTN7 | .631 |
VHBD1 | .602 |
VHBD2 | .586 |
VHBD3 | .535 |
VHBD4 | .615 |
VHBD5 | .580 |
VHBD6 | .481 |